New Resources for Norovirus
Jennifer Chang, Jover Lee, Kim Andrews, James Hadfield, Allison Li, Katie Kistler, John Huddleston, Trevor Bedford
Historically called "winter vomiting disease" (Adler and Zickl, 1969), norovirus is the bane of any parent, long term care facility, or otherwise contained community. The name of the virus is a mangled reference to Norwalk, Ohio, where a particular 1968 outbreak surged through a population of school children within a 24 hour period. Despite its explosive symptomology, the etiological agent was not identified as a virus until 1972, by immune electron microscopy (Kapikan et al, 1972) and not genetically sequenced until 1989 (Jiang et al, 1993). The subsequent classification of the virus was fraught as it became clear that the virus undergoes recombination, frequently between the ORF1 and ORF2 region (Bull et al, 2005), resulting in different evolutionary histories between the polymerase (RdRp in ORF1) and surface capsid (VP1 in ORF2). Despite multiple attempts and discomfiture among those afflicted, there is no approved norovirus vaccine.
# Phylogenetic analysis
Nextstrain provides regularly updated phylogenetic monitoring of norovirus along several different facets. Since this is a highly recombining virus, we provide individual gene trees as well as the full genome tree of all norovirus samples, building off the prior effort of Allison Li and Katie Kistler and John Huddleston. They have also faceted the genome trees along important VP1 types GII.2, GII.3, GII.4, GII.6 and GII.17. Therefore, as of the time of this writing, we provide 14 regularly updated views of norovirus evolution.
| group | genome | p48 | NTPase | p22 | VPg | 3CLpro | RdRp | VP1 | VP2 |
|---|---|---|---|---|---|---|---|---|---|
| all | genome | p48 | NTPase | p22 | VPg | 3CLpro | RdRp | VP1 | VP2 |
| GII.2 | genome | ||||||||
| GII.3 | genome | ||||||||
| GII.4 | genome | ||||||||
| GII.6 | genome | ||||||||
| GII.17 | genome |
The combination of highly diverged and recombined sequences proved a challenge in rooting the phylogenetic trees, and we advise that any results should be interpreted with caution. Even so, the Nextstrain trees are provided as a broad summary of the genetic diversity and relatedness, and further biological interpretation may require targeted sampling, tuning of parameters, a different alignment reference, or focusing on particular gene combinations. The trees are being annotated by both VP1 and RdRp types. From the map, norovirus types do not appear to be geographically segregated. From the frequency panel, we see indications that there are dynamics of leading types and it is not a virus that has reached genetic equilibrium of the proportion of those types.
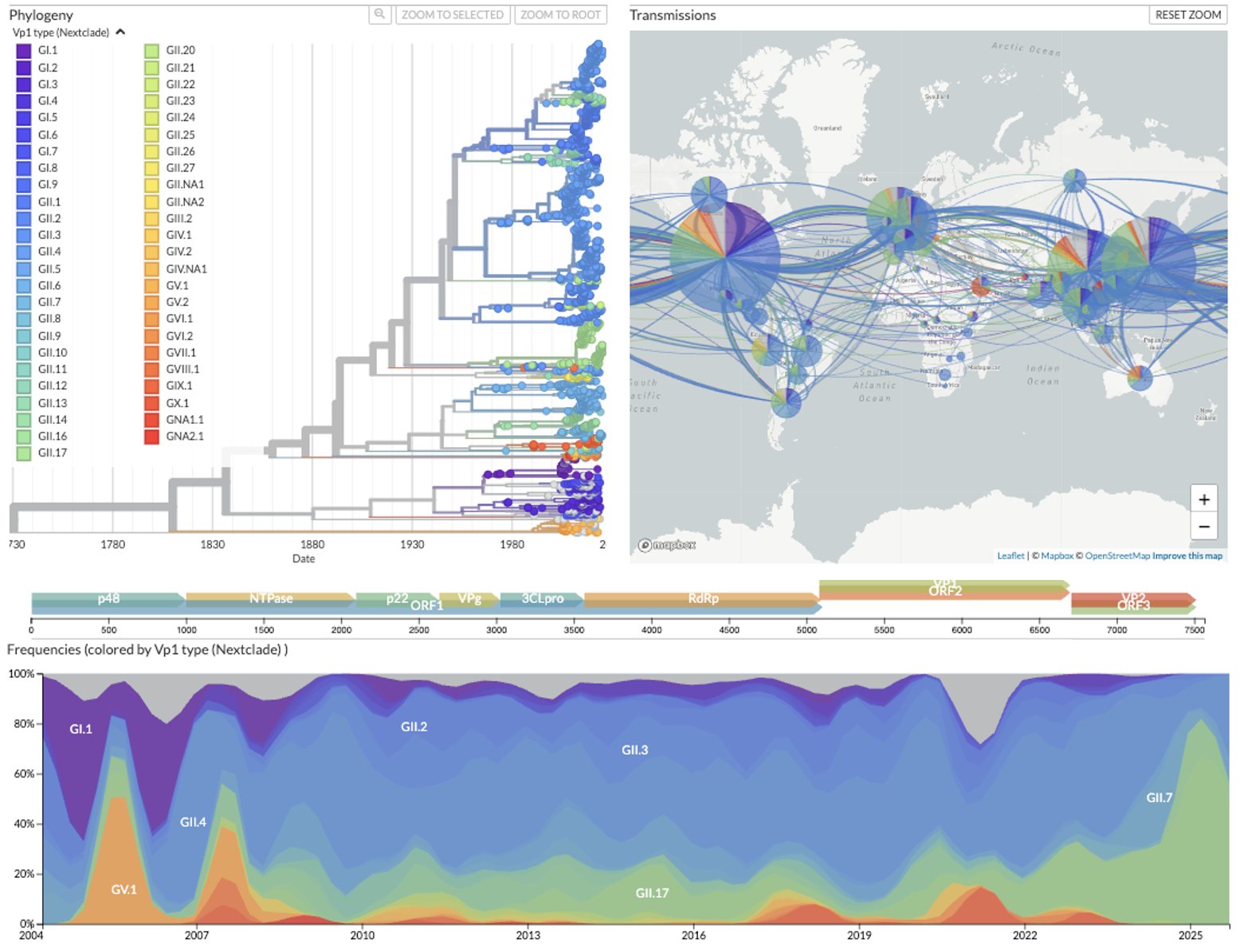 Figure 1. Norovirus is globally distributed and highly divergent. Phylogenetic trees are annotated by both VP1 and RdRp types, host, country, date, genome and gene coverage percentages. The full genome tree is shown here. From the map, norovirus types do not appear to be geographically segregated. From the frequency panel, we see indications that there are dynamics of leading types and it is not a virus that has reached genetic equilibrium of the proportion of those types.
Figure 1. Norovirus is globally distributed and highly divergent. Phylogenetic trees are annotated by both VP1 and RdRp types, host, country, date, genome and gene coverage percentages. The full genome tree is shown here. From the map, norovirus types do not appear to be geographically segregated. From the frequency panel, we see indications that there are dynamics of leading types and it is not a virus that has reached genetic equilibrium of the proportion of those types.
# Norovirus groups, types, and variants
Norovirus samples have a dual-typing system based on a polymerase region (RdRp) and capsid region (VP1) of the genome, between which is a known recombination site. The resolution of norovirus typing has undergone multiple changes (Zheng et al., 2006; Eden et al., 2013; Chhabra et al., 2019; Tatusov et al., 2020), but generally are split into a "genogroup", "genotype", and "variant" classification for VP1, and "P-group", "P-type", and "variant" for RdRp. For the sake of naming Nextstrain trees, we will name these VP1 group, type, or variants and RdRp group, type or variants respectively.
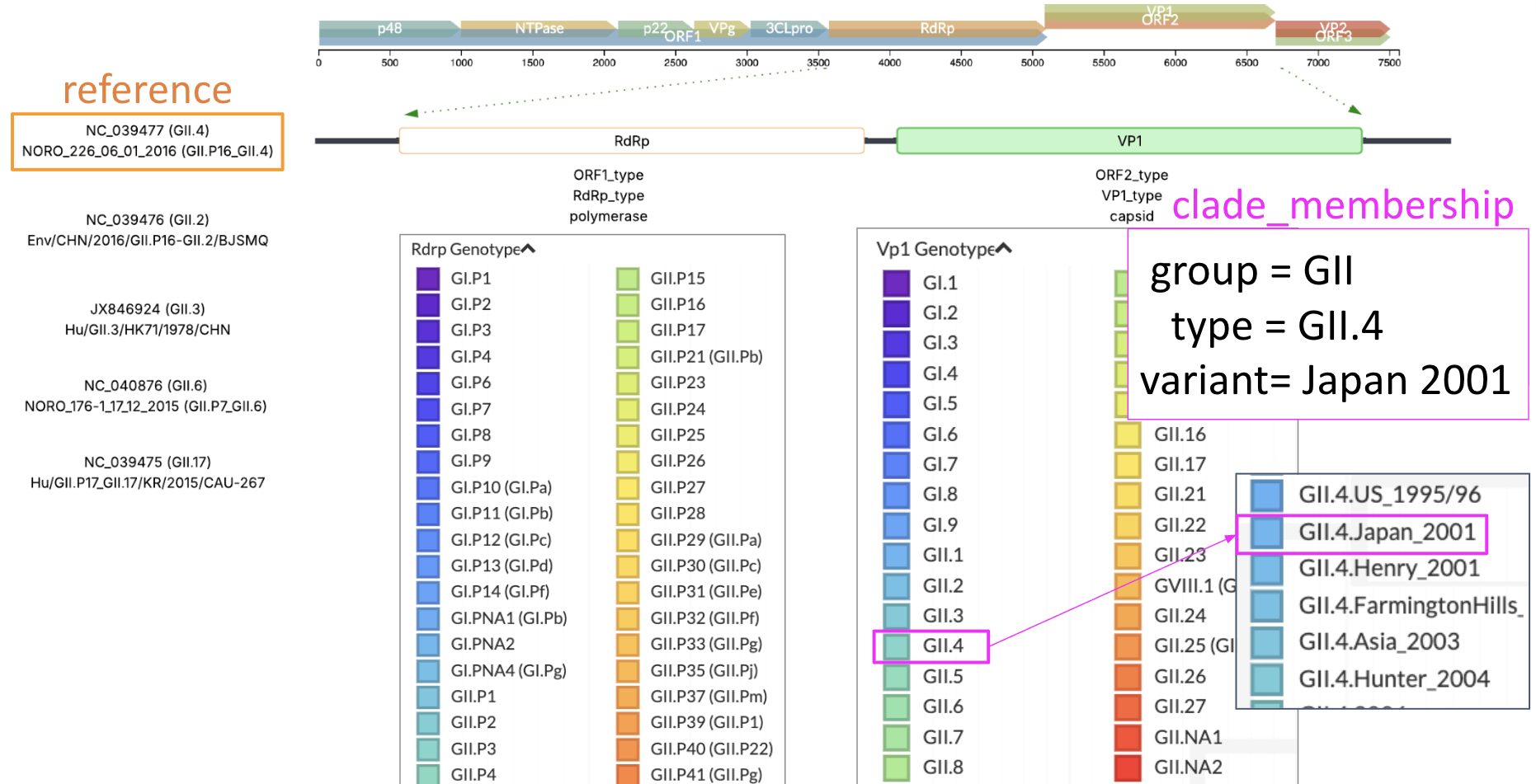 Figure 2. Typing of norovirus samples is based on the VP1 and RdRp region and are further split out into group, type, and variant resolution.
Figure 2. Typing of norovirus samples is based on the VP1 and RdRp region and are further split out into group, type, and variant resolution.
Group, type, and variant levels of resolution were roughly classified by a preliminary Nextclade datasets based on either VP1 or RdRp gene. The Norovirus nextclade datasets are preliminary and further development is pending. These datasets have been built from scaffold strains listed at the Human Calicivirus Typing Tool as of September 16, 2025.
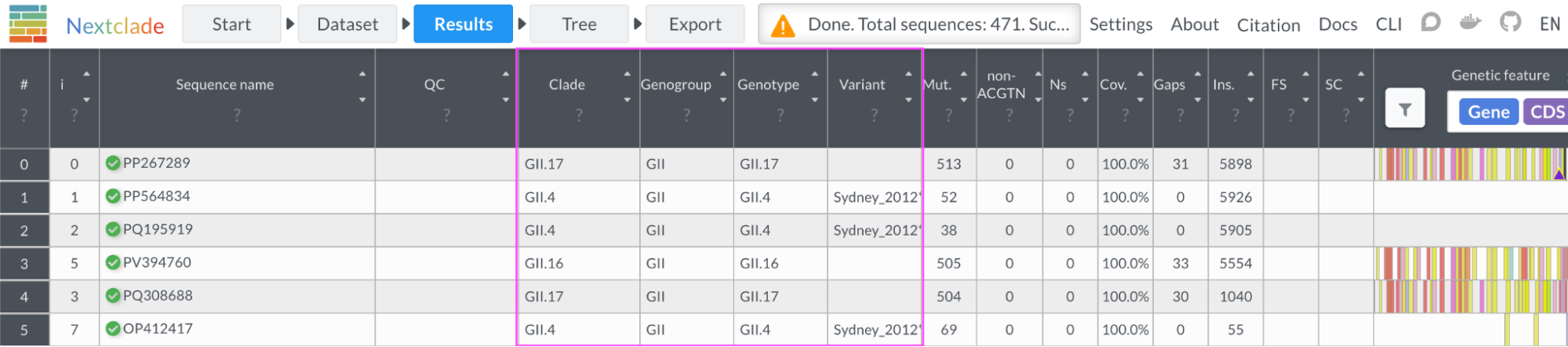 Figure 3. Preliminary norovirus classification into Group, Type, and Variant columns.
Figure 3. Preliminary norovirus classification into Group, Type, and Variant columns.
# Nextstrain resources
We curate sequence data and metadata from NCBI as the starting point for our analyses. We provide snapshots of the exact curated sequences and metadata for norovirus workflows at:
# Acknowledgments & request for comments
We welcome comments or suggestions from norovirus researchers to improve these Nextstrain and Nextclade datasets. Special thanks for feedback from Chao-Yang Pan and Erik Wolfsohn for answering questions and providing some biological context.